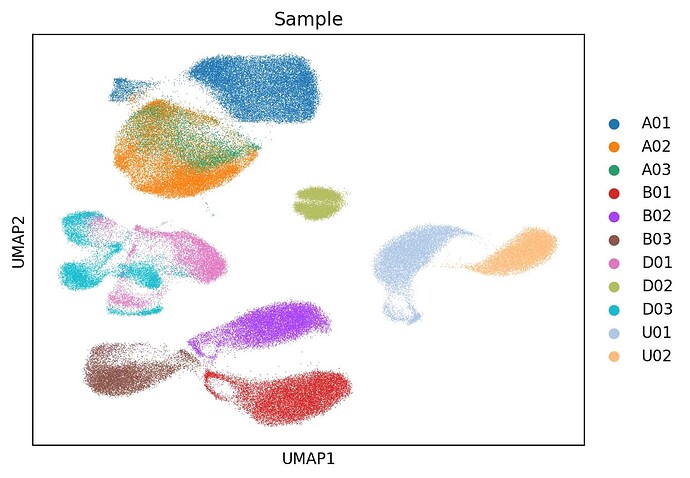
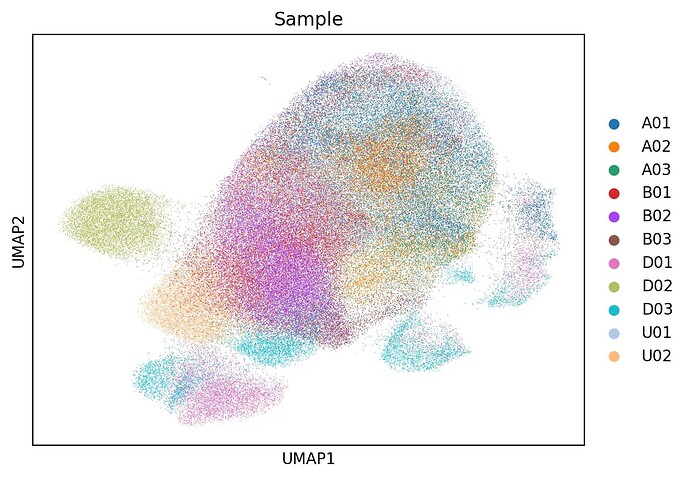
Dear Community,
I’m currently working on integrating a single-cell RNA-seq dataset of human mesenchymal stem cells (MSCs) using scvi-tools. The dataset includes 11 samples, each from a different donor, across four tissue types:
- A: Adipose (A01–A03)
- B: Bone marrow (B01–B03)
- D: Dermis (D01–D03)
- U: Umbilical cord (U01–U02)
Each sample corresponds to one patient, so I’ve been using the sample ID (e.g., A01, B02) as the batch_key in SCVI.setup_anndata.
My goal is to mitigate donor-specific batch effects within each tissue, but preserve the biological differences between tissues (since tissue-of-origin is an important axis of variation here).
I’ve followed the scvi-tools tutorials, but after integration, the tissue-specific structure seems to be partially lost.
My Questions:
- Is using
batch_key='Sample'the right approach here? - Should I treat tissue type as a
categorical_covariateinstead, to help scVI retain inter-organ differences? - Has anyone dealt with a similar situation where batch effects should be removed within groups but preserved between groups?
Any advice or best practices for this type of integration would be greatly appreciated!
Thanks in advance!