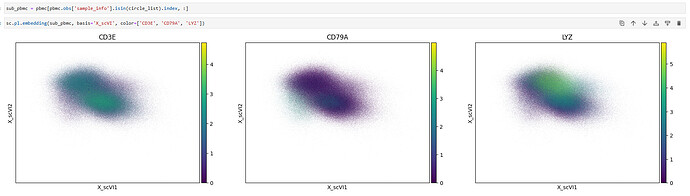
When using scVI for batch correction and clustering on my single-cell data, I observed an unusual result: all cells from the same sample are clustered in a circle, separated from the rest. Changes to n_latent , training epochs, and HVG numbers did not resolve the circular artifact. The UMAP plot is attached for reference (see cluster 10)
Below are my codes:
import scanpy as sc
import torch
import seaborn as sns
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
import random
from collections import defaultdict
torch.set_float32_matmul_precision("high")
pbmc = sc.read_h5ad('/data/work/data/raw_data/ESCC_PBMC_All_Basic_QC.h5ad')
# downsample each sample for hvg calculation
sample_key = 'sample_info'
n_subsample = 5000
pbmc_list = []
for sample in pbmc.obs[sample_key].unique():
pbmc_sample = pbmc[pbmc.obs[sample_key] == sample].copy()
n_cells = pbmc_sample.shape[0]
if n_cells > n_subsample:
idx = np.random.choice(n_cells, size=n_subsample, replace=False)
pbmc_sub = pbmc_sample[idx, :].copy()
pbmc_list.append(pbmc_sub)
else:
pbmc_list.append(pbmc_sample)
pbmc_meger = sc.concat(pbmc_list, label='tmp_info')
sc.pp.highly_variable_genes(
pbmc_meger,
flavor="seurat_v3",
n_top_genes=3000,
batch_key="tmp_info",
span=0.8
)
hvg_genes = pbmc_meger.var[pbmc_meger.var['highly_variable']].index.tolist()
pbmc.var['highly_variable'] = pbmc.var_names.isin(hvg_genes)
sc.pp.normalize_total(pbmc, target_sum=1e4)
sc.pp.log1p(pbmc)
pbmc.raw = pbmc
import scvi
import tqdm as notebook_tqdm
scvi.model.SCVI.setup_anndata(pbmc, layer="counts", batch_key="sample_info",
continuous_covariate_keys=['doublet_score', 'n_genes_by_counts'],
categorical_covariate_keys=['patient_num'])
model = scvi.model.SCVI(pbmc, n_layers=2, n_latent=30, gene_likelihood="nb")
model.train(batch_size=512, max_epochs=32)
latent = model.get_latent_representation()
pbmc.obsm['X_scVI'] = latent
latent.shape
sc.pp.neighbors(pbmc, use_rep='X_scVI', random_state=26)
sc.tl.leiden(pbmc, flavor='igraph', n_iterations=-1, resolution=0.3)
sc.tl.umap(pbmc)
sc.pl.umap(pbmc, color='leiden')
There is no explicit error output, but the UMAP result is abnormal (see attached image).
scVI Versions: 1.1.6.post2
as shown in elbo_train and train_loss_epoch figures, the model had learned the data feature.