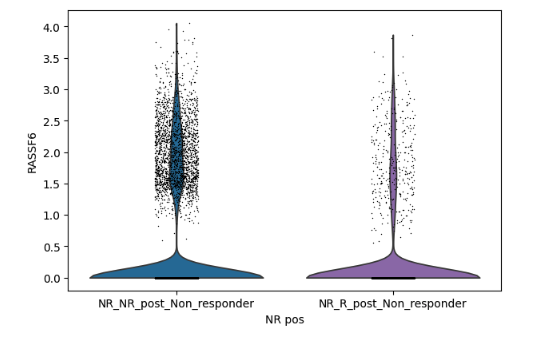
Hello, after using model.differential_expression(), for a specific gene of interest I get the following outcome:
proba_de 0.9724
proba_not_de 0.0276
bayes_factor 3.561951
scale1 0.000764
scale2 0.000962
pseudocounts 0.0
delta 0.15
lfc_mean 0.612965
lfc_median 0.933985
lfc_std 2.177583
lfc_min -5.728889
lfc_max 6.05063
raw_mean1 0.4188
raw_mean2 0.502647
non_zeros_proportion1 0.1432
non_zeros_proportion2 0.234677
raw_normalized_mean1 3.688732
raw_normalized_mean2 8.833171
is_de_fdr_0.05 True
How can it be that the lfc_mean is positive (0.61), whereas the gene is clearly more expressed in the second group?