Hi all,
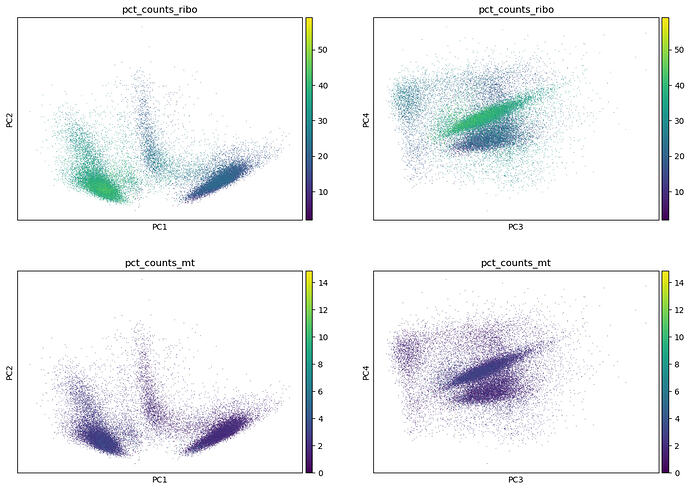
Following the tutorial of scanpy about clustering ( Preprocessing and clustering — scanpy ), I plotted some QC metrics (computed with sc.pp.calculate_qc_metrics(adata, qc_vars=[“mt”, “ribo”])) on the first four PC. The goal is to see if some undesired features drive variation in my data. And indeed, I see that the ribosomal genes show patterns of expression along some PC axis (cf screenshot).
I wonder if these genes should be removed then? Because we always hear about the mitochondrial genes to be removed but never about the ribosomal genes.
I am working on a human head sample taken at stage week 8 post-conception.
Thank you for your precious insights ![]()